
Our research aims to address two broad questions in developmental biology: How are tissue size and shape precisely controlled during early mammalian development? How is tissue geometry sensed and transmitted to the cellular level to impact cellular functions? How are mechanics and biochemical signalling integrated across multiple scales to ensure robust morphogenesis and patterning?
To address these fundamental questions, we focus on understanding mammalian follicle development, which is critical for the maturation of functional eggs for successful reproduction. We aim to identify the mechanobiology principles underlying mammalian follicle growth, and decipher the roles of tissue mechanics and fluid forces in oocyte quality control, using ex vivo mouse ovaries and follicle culture. A quantitative and mechanical understanding of folliculogenesis will deepen our understanding of reproductive biology and ageing, with important implications for assisted reproductive technology (ART), infertility treatment and mechano-therapeutics.
Proteins that are activated or suppressed by mechanical force allow eukaryotic cells, such as those that make up the human body, to respond to changes in their microenvironment. This is crucial during development, when cell niches are still forming. Similarly, the changes in our bodies that occur as we age can lead to an altered physical environment for our cells—for example, bones may become softer, and scar tissue can build up.
By manipulating specific proteins and subjecting cells to a level of physical force consistent with that which occurs naturally in the human body, we are gaining a better understanding of how various molecular machines detect and transduce physical forces. Of particular interest are contractile cytoskeletal units, cadherin-based adhesion complexes, which connect cells with one another, and integrin-based adhesion complexes, which enable cellular interaction with the extracellular matrix.
Several proteins found in these machines can be stretched, thereby exposing domains that will bind additional proteins. This mechanism is replicated in MBI laboratories at the level of single proteins. Understanding the forces required to stretch a protein, to trigger contraction of an actomyosin based complex, or to induce a biochemical change in a protein provides crucial information on how cells integrate physical force into biochemical pathways.
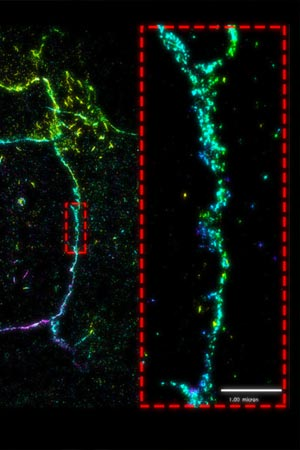
superresolution imaging reveals that E-cadherin assembles as distinct, punctate clusters. Detailed imaging at the nanoscale level demonstrates that these clusters do not merge (dotted red box). Read the full article: Defining Adhesion Clusters
Signals established at the cell periphery, or within the cytoskeleton, may converge on the cell nucleus where they will impact genome regulation and protein expression.
At the MBI, we investigate how mechanical cues regulate key signaling pathways, such as the Hippo pathway, and subsequently, the activity of various transcription machinery. In these cases physical cues, which may generated through the contraction or stretching of a protein complex, can lead to the recruitment or activation of other enzymes and transcription factors. Physical force is subsequently converted to a biochemical signal.
Alternatively, mechanical signals may be transferred through the cell as a physical force. For example, when the cytoskeleton contracts, or when an alteration in cell morphology generates tension within the nuclear membrane, the forces involved are transferred throughout the cell. The effect of transferring force through structural elements of the cell is investigated at the MBI. For example, we grow cells on substrates of a defined shape and monitor how nuclear morphology is affected, and in turn, how this phenomenon alters DNA packaging and the spatial arrangement of genes. Through this work, the importance of mechanics in the regulation of the genome is becoming evident.
Like their larger multicellular hosts, bacteria and other prokaryotic cells are subjected to, and generate, mechanical forces. When invading a host cell, forces must be overcome, and in some cases, the dynamic structures of the host cell may be exploited by bacteria to facilitate their invasion.
For example, bacteria can modulate the cell membrane and manipulate the cytoskeleton for entry into the host, or for the secretion of virulence factors. In doing so the bacteria are able to establish an infection. However, the precise molecular mechanisms underlying bacterial uptake, and the mechanisms by which bacteria can evade the body’s defenses remain unclear. At MBI, we are investigating the biophysical aspects of bacterial pathogenesis, and in particular, are interested in the mechanisms by which bacteria like Salmonella utilize the endocytic membrane trafficking pathway to survive within the host. In this case, Salmonella will survive within an endosomal compartment called the salmonella containing vacuole (SCV). Salmonella also thrives within its host in a non-infectious phase for an extended period of time. Here, the bacteria will exist as a biofilm. How salmonella lives within SCVs, the various cytoskeletal modifications that take place during invasion and the regulatory mechanisms that are modulated to enable Salmonella to exist as a biofilm, are all researched at MBI.
Additional work in this area is looking at the formation and nature of cytoskeletal pedestals, which, in the case Enteropathogenic E.coli (EPEC) infection, are found within host cells at the site of bacterial attachment.
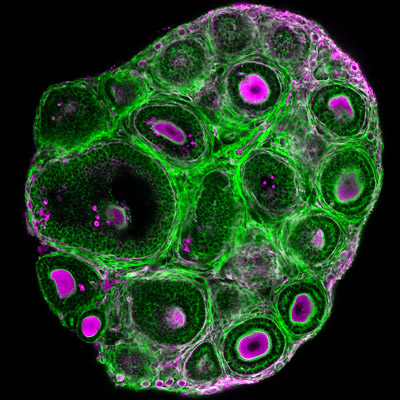
While the follicular stages have been well characterized by histological criteria, the mechanisms underlying follicular growth remain poorly understood, partly due to a lack of understanding of the mechanical interactions between the oocyte and its surrounding somatic cells and extracellular matrix (ECM). We will develop novel biophysical tools to measure and study these mechanical processes. In combination with deep tissue imaging and physical and genetic manipulations, we aim to unravel key mechano-signaling pathways controlling follicle morphogenesis and the oocyte quality, as proposed in our recent review (Front. Cell Dev. Biol. 2022). One approach is Brillouin microscopy, which we have shown to reveal mechanical compartmentalization during follicle and embryo maturation (Comm. Biol. 2021, Nat. Methods 2023).
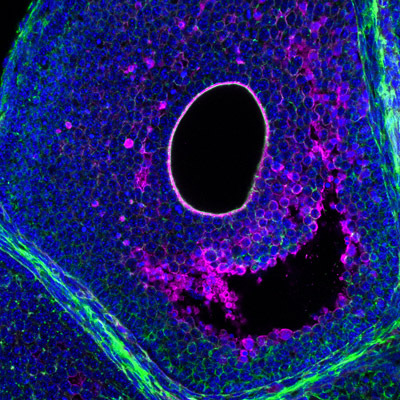
An important, albeit understudied aspect is the role of tissue hydraulics in development and reproduction (see our review in Semin. Cell Dev. Biol. 2022). We have recently shown that luminal pressure and signalling play a critical role in regulating the tissue size and cell fate specification of mouse blastocysts (Nature 2019, Dev. Cell 2019), and proposed a unifying framework to understand the interplay between luminal mechanics, signaling and cell fate during tissue morphogenesis (Development 2020). We have also worked with others to reveal the mechano-osmotic control of intestinal organoid morphogenesis (Nat. Cell Biology 2021). Interestingly, during mammalian folliculogenesis, a similar process occurs where a fluid-filled cavity emerges at the antral follicle stage. We will study the dynamics and mechanisms of luminogenesis, and investigate how luminal pressure and signalling collectively influence the oocyte development, using deep tissue imaging, machine learning, biophysical modelling and genetics.

Boon Heng joins the Chan Lab as a PhD student, having obtained his B.Sc. in Life Sciences from the National University of Singapore in 2021. His previous research spans from C. elegans embryogenesis to zebrafish somite formation. Using a mouse model, he wishes to explore how mechanics and biochemical signalling are tightly coordinated to drive robust tissue patterning in pre-antral follicle development. Learn more

Boon Heng joins the Chan Lab as a PhD student, having obtained his B.Sc. in Life Sciences from the National University of Singapore in 2021. His previous research spans from C. elegans embryogenesis to zebrafish somite formation. Using a mouse model, he wishes to explore how mechanics and biochemical signalling are tightly coordinated to drive robust tissue patterning in pre-antral follicle development. Learn more

